When a mobile phone takes a picture, software organizes the tiny pixels into an image that humans can see. But when an international team of researchers captures information at the genomic level in cells, it takes help from engineers at the Ohio University Bioinformatics Lab to organize the data and find patterns in the 3D picture that lets scientists visualize the genes at work.
With help from Lonnie Welch, a team led by Ana Pombo at the Max Delbrück Center for Molecular Medicine in Germany has deciphered the 3D structure of the genome in three cell types of the mouse brain. Their continuing research characterizing complex genomic interactions will provide insights about neurological disease, said Welch, the Stuckey Professor of Electrical Engineering and Computer Science in the Russ College of Engineering and Technology.
“What happens in the brain when we learn something new? What happens when we develop an addiction or a neurological disease? Which genes spring into action in the brain cells at such moments? And how is the whole process regulated in the cell nucleus?” These sorts of questions have been driving the team’s work for many years, reports the Berlin-based Max Delbrück Center for Molecular Medicine in the Helmholtz Association in describing the impact of their research.
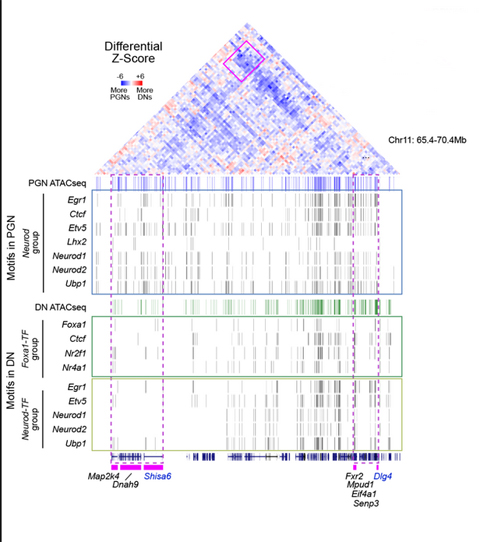
Pombo, Welch and the team recently published an article in the journal Nature where they report discoveries about the genome not previously seen by other approaches. They discuss their discovery of very long genes that almost completely unfold their structure when they are “at work,” as well as specific folding patterns and a phenomenon that Welch calls “word pairs.” The word pairs are a pattern identified by the OHIO researchers.
The small set of genomic word pairs that appear within the neuron-specific genomic contacts “bring us closer to understanding the underlying molecular mechanisms, which we plan to study next,” said co-author Alexander Kukalev.
Discovering new folding patterns in genes
“After the human genome was decoded, it has become clear that only a very small part of the DNA actually contains the instructions for making proteins,” Pombo says. The other much larger part, she says, was initially considered superfluous and often dubbed junk DNA. “Yet it turns out that these mysterious regions of the genome contain the crucial information that regulates the activity of genes by direct spatial contacts with them or by influencing their position,” Pombo said.
“To learn more about these tiny switches that turn genes ‘on’ and ‘off,’ Pombo and her team developed a technique called genome architecture mapping, or GAM for short, which they described in Nature in 2017. The technique creates 3D maps of the entire genome and uses statistical calculations to determine which sections of the genome prefer to connect with each other – in other words, the locations of the switches that regulate each gene,” reports the Max Delbrück Center.
In their latest study, they use the GAM technique in combination with immunoselection, where they label three different cell types from the mouse brain, to study 3D chromatin structure of both the active and the inactive DNA.
The first co-author, Warren Winick-Ng notes that “a major advantage of combining immunoselection with the GAM technique is that we can study all cells in their native tissue environment without having to remove them from their tissue context.”
Next step in neurological disease research
Welch and the OHIO Bioinformatics Lab team’s contributions to this research included computational 3D genomics, focusing on the discovery of patterns that characterize complex genomic interactions. The lab, which now includes four graduate students and three undergraduates, developed all the necessary data mining software for this research, which they deployed on a computer cluster in the college’s School of Electrical Engineering and Computer Science.
“Our recently published joint study employed GAM to assess the 3D genome structure in two types of neurons and in glial cells. With the help of Yingnan Zhang (OHIO Electrical Engineering and Computer Science Ph.D. student), we discovered that the folding of the DNA strands and the contacts between the individual genomic elements are each highly specific in different types of brain cells. OHIO Bioinformatics developed novel supervised machine learning algorithms to search for patterns in interacting genomic regions, resulting in discovery of a small set of genomic ‘word pairs’ that appear within the neuron-specific contacts and may serve as underlying molecular mechanisms of the 3D genome,” Welch said.
With funding from the NIH 4D Nucleome Program and the Charité Neurocure Center of Excellence, the collaborators at the Berlin Institute for Medical Systems Biology have begun mapping the 3D genomes of human individuals and patients with neurological disorders.
“Our results provide a foundation for further study of neurology and of nuclear organization,” Welch said, adding that the international team is now analyzing brain tissue from deceased patients with amyotrophic lateral sclerosis (ALS), as a new collaboration with Berlin’s Charité research hospital.
“We are expanding our computational methods to enable analyses of this new 3D genome data. Specifically, we are developing computational capabilities for studying the multiway chromatin contacts uniquely captured by GAM. This will create the first algorithms for both discovering and characterizing complex (n-way) genomic interactions derived with the GAM technique. These methods will enable deep understanding of high-order 3D-genomic interactions and will yield new insights into the mechanisms and pathologies of neurological diseases.”
The lab’s work featured in the Nature paper was funded in part by a grant from the Ohio University Graduate Education and Research Board. Based on his work on this project, Welch was accepted into the National Institute of Health’s 4D Nucleome (4DN) Consortium as an Associate Member in February of 2022.
Welch is also a member of the Molecular and Cellular Biology program at OHIO, director of the Bioinformatics Laboratory at Ohio University, and the founder and Chair of the Great Lakes Bioinformatics Conference, an official conference of the International Society on Computational Biology. His research has been sponsored by the National Human Genome Research Institute, the Ohio Plant Biotechnology Consortium, NASA, the National Science Foundation, the Defense Advanced Research Projects Agency, and the Ohio Board of Regents. He was the principal investigator of the $9M Bioinformatics Program of the Ohio Board of Regents and Director of OHIO’s Bioinformatics Certificate Program. Welch is founder and co-chair of the International Society on Computational Biology’s Special Interest Group on Regulatory and Systems Genomics (RegSys).




















Comments