By Amanda Biederman
NQPI editorial intern
The human body is home to more than 200 different types of cells, each performing a unique function. But what makes a blood cell a blood cell, or a neuron a neuron?
All cells in an organism are derived from a single embryonic stem cell. Cells exhibit different patterns in their gene expression throughout the life of the organism. This allows for cellular reprogramming, modifying the gene expression to convert one cell type into another. This offers potential for therapeutics in numerous diseases such as cancer and neurodegenerative diseases.
Physics & Astronomy Professor and Nanoscale and Quantum Phenomena Institute member Dr. Horacio Castillo and recent alumnus Dr. Sai Teja Pusuluri have developed a novel approach to characterize cell differentiation. Working with Boston University professor Dr. Pankaj Mehta and BU alumnus Dr. Alex Lang, they simulated a computational model to describe patterns of gene expression in mouse fibroblast cells.
For decades, scientists have described the process of cell differentiation using the “epigenetic landscape” image, which visualizes a differentiating cell as a sphere that travels across a landscape and eventually settles into a “valley,” representing a differentiated cell type.
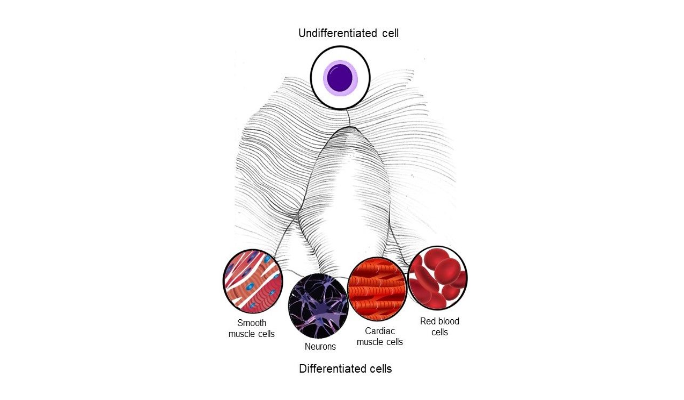
According to Waddington’s epigenetic landscape, cell differentiation is determined by the movement of a cell through a series of paths that lead to basins, representing its possible mature forms. (Image adapted from Wikimedia Commons)
“The landscape has been in principle just an image,” Castillo said. “(But our work) seems to indicate that a model based on a landscape actually can be used to describe how these genes work.”
Castillo’s group implemented the landscape model into their approach, converting a classical metaphor into a quantitative tool. They constructed a spin-glass model, which fits specific patterns of gene expression to a discrete cell type. Spin-glass models have been used previously to describe neural networks. With this model, the group described how landscapes govern cell differentiation.
The group is now characterizing the correlation structure of gene expression patterns among different cell types. They are constructing random trees to visualize the hierarchy of cell types, with embryonic stem cells as the root and differentiated cells as the branches. With this approach, Castillo said, the group is working to better understand the dynamics of this complex, yet fundamental phenomenon.




















Comments